Data Mining of Protein Sequences with Amino Acid Position-Based Feature Encoding Technique
Novel Amino Acid Position-Based Encoding for Protein Sequence Classification
Abstract.
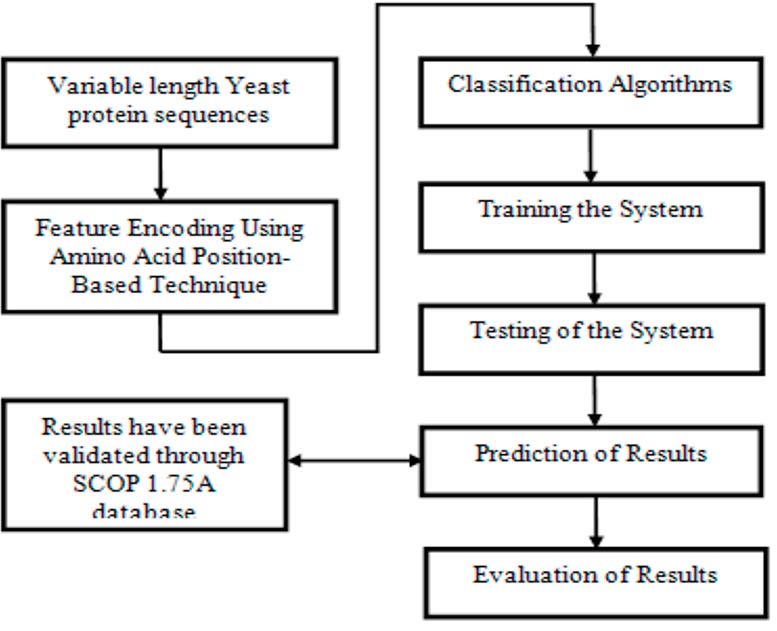
We propose a novel amino acid position-based feature encoding technique for the classification of protein sequences, achieving a significant improvement in classification accuracy. Each amino acid’s occurrence positions in a sequence are encoded into a fixed-length numeric feature vector, where the mean \(\mu_{i} = \frac{1}{n} \sum_{j=1}^{n} p_{ij}\) and variance \(\sigma_{i}^{2} = \frac{1}{n} \sum_{j=1}^{n} (p_{ij} - \mu_{i})^2\) are computed for each amino acid. Our experiments on Yeast protein sequences using a decision tree classifier yielded a classification accuracy of 85.9%, outperforming previous methods. The proposed technique efficiently encodes sequence data, enabling accurate predictions of protein structure and function from primary sequences. This methodology demonstrates robustness, reliability, and high accuracy, providing a significant advancement in the computational biology domain.
Illustration of the proposed experimental design.