Efficient Feature Selection and Classification of Protein Sequence Data in Bioinformatics
Enhanced Accuracy in Protein Classification via Statistical Metric-Based Feature Selection
Abstract.
Bioinformatics requires efficient classification of protein sequences to predict their structure and function. We propose a statistical metric-based feature selection technique combined with an n-gram encoding method to improve classification accuracy. Protein sequences are encoded using descriptors of lengths 1, 2, and 3 amino acids, resulting in a feature vector space \(\mathbf{X} = \{X_1, X_2, \ldots, X_{8420}\}\). The statistical metric for feature selection is defined as \(V_d (j) = \min_{p \neq q} \left\{ \frac{|X_p(j) - X_q(j)|}{\sqrt{\frac{S_p^2(j)}{N_{\text{Total}}} + \frac{S_q^2(j)}{N_{\text{Total}}}}} \right\}\), where \(S^2_i(j)\) represents the variance of the \(j\)-th feature in the \(i\)-th superfamily. This technique reduces feature dimensionality and enhances classification accuracy by approximately 15% to 20% across various datasets, outperforming traditional alignment-based methods. Experimental results on datasets from the UniProt database demonstrate the effectiveness of our approach, achieving an accuracy of up to 96% with neural network classifiers.
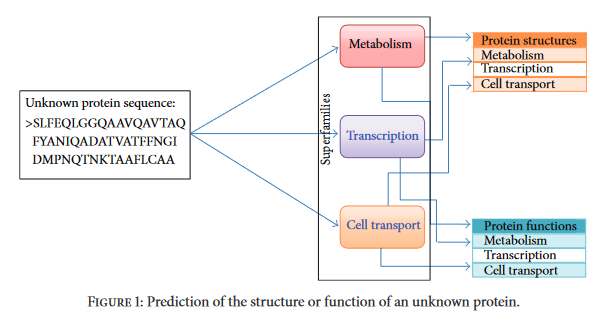
Illustration of the proposed experimental design.